 |
 |
|
 |
Interview with Jack Johnson
recorded and edited by Sondra Schlesinger
November 5, 2000
St. Louis, MO
John (Jack) Johnson is a Professor in the Department of Molecular Biology at The Scripps Research Institute, La Jolla, CA. It doesn't take long to catch Jack's enthusiasm for viruses. I know how wonderful and exciting viruses are, but, if I didn't, Jack would have convinced me of their fascination in only a few minutes. Jack started his scientific career in inorganic physical chemistry and, as he told me, there was a time when the term icosahedral symmetry was the only phrase about viruses that he understood. But that was in the distant past and now he is deeply involved in trying to understand the dynamics of virus structure and its importance in the infectious process. Jack's history in virology begins in 1972 when he joined Michael Rossmann's lab and in many ways parallels the theme of Viruses from Structure to Biology. He told me some of that history in our interview.
SS Jack, you were a graduate student in chemistry, what made you think about going to Michael (Rossmann) for postdoctoral training?
JJ The circumstances were very difficult in 1972. My Ph.D. was in physical and inorganic chemistry which I enjoyed very much and I really intended to develop my career in that direction but there were no job opportunities. There were no postdoctoral fellowships in that area, there were no industrial jobs in that area. I was really faced with a decision of either leaving science altogether or making a dramatic change in the direction of my career. I was very fortunate in that I worked with Robert Jacobson at Iowa State University. Bob was a graduate student with Bill Lipscomb, when Michael Rossmann was a postdoc in Lipscomb's lab. Scott Mathhews was also a graduate student with Lipscomb at that time. Bob suggested that I consider changing directions and going into structural biology or protein crystallography since there were opportunities in those areas. He told me to write to both Michael and Scott (Mathews). Michael quickly responded and offered me a postdoctoral position, presumably on the recommendation of Bob Jacobson because we hadn't met or talked or anything. So that's how I got into protein crystallography and virus crystallography.
SS So you had no particular foresight that viruses would be interesting?
JJ No, in fact, ultimately when I chose the project I was going to work on with Michael, the only thing that I was familiar with, in terms of the opportunities in Michael's lab, was icosahedral symmetry because I had done molecular orbital calculations on boron hydrides as a graduate student. Michael, at that time, was interested primarily in enzymes, lactate dehydrogenase, glyceraldehyde-3 phosphate dehydrogenase. He mentioned at the end of our interview that a project that he wanted to get started in the lab was virus crystallography. When he said that viruses had icosahedral symmetry, it was the first words I really understood in the conversation and that motivated me to go in that direction.
SS But he said that you were crazy to choose that subject.
JJ I wasn't crazy, I was desperate, so a desperate man does crazy
things, I guess. It turned out to be a wonderful, serendipitous set of circumstances
that pushed me in that direction.
This is what Michael Rossmann remembers about Jack's first visit:
"I had just obtained a diffraction picture of a CCMV crystal. This was my first diffraction
from a virus crystal at Purdue, although I had worked with excellent STNV crystals in Bror
Strandberg's lab while I was on sabbatical there in '71. I was overjoyed to get a start with
virus crystals at Purdue although they probably diffracted to only about 15 angstroms
resolution. I showed these pictures (I think they were still wet in the dark room) to Jack.
Much later Jack told me that he could not understand why these pictures were so special,
but he did his best to be interested. Jack has come a very long way from those beginnings!
SS So you went to Michael's lab in 1972.
JJ September '72
SS What was it like to go from what was straight chemistry to biology? I assume you didn't have to grow the virus.
JJ Actually, I did grow virus. Michael had a technician in the lab, Mary Ann Wagner, who had started to grow plants and inoculate them with virus and I was really quite fortunate because Andy Jackson ( Andy is a plant virologist now at the University of California, Berkeley) was at Purdue at that time. He was a brand, new assistant professor and sympathetic to people that were trying to do things that they didn't know how to do. Andy and Richard Lester were quite instrumental in helping me get started in the world of plant virology. Mary Ann, also a plant virologist, and I prepared virus and sprayed green sap all over each other when the bags that we were using for separation would break. Yes, it was quite a new experience for me, something that I really embraced and was excited about right from the beginning.
SS Was southern bean mosaic virus the one that you started with?
JJ No, we started with cowpea chlorotic mottle virus (CCMV). This was a kind of an historical development from Michael's relationship with John Bancroft who was at Purdue until about 1971. He had left by the time I arrived, but John had done a wide variety of assembly studies with CCMV and then we tried to grow crystals of that but those crystals didn't diffract X-rays. Because of work that Beatrice Magdoff had published many years earlier on the crystallization of southern bean mosaic virus, we thought we would give that a try and that indeed turned out to be the one we could get good diffracting crystals.
SS You started in '72, when did you get crystals?.
JJ We had crystals fairly early. It didn't take more than a few months. Ira Smiley was a postdoc in Michael's lab who had set up some crystals. I think we were probably getting our first diffraction patterns by early '73. We had no rotating anode X-ray generator. We had only a sealed tube X-ray generator and all we could take were what are called still photographs so you could only get patterns that would tell you what the lattice constants of the crystals were. You couldn't actually record amplitudes from which you could get structural information. The first year or so that I spent at Purdue, it was strictly characterizing the lattices of these crystals without being able to get any direct structural information. But even that was interesting because the lattice told you how the particles were packed and I quickly could use some of my inorganic experience with crystalline elements. If you look at crystals of elements, they pack in very simple patterns because basically they are spheres and the viruses are doing the same thing: they were packing in simple patterns because they are spherical. So throughout my career I have reached back into my inorganic days to help me understand, maybe in ways protein crystallographers might not have, how some of these crystallographic issues could be dealt with.
SS Did you go to the Cold Spring Harbor meeting on protein crystallography?
JJ The famous 1971 Cold Spring Harbor meeting. No, but the echoes of that exchange were still very much taking place in the laboratory. I remember Michael asking me to read the rebuttal manuscript that he wrote to Aaron Klug's rebuttal to Michael and Bror Strandberg's paper at Cold Spring Harbor. And this was quite a time. This was my first realization that I had stumbled into a very high profile laboratory. I knew it was a strong lab, but suddenly there were all these controversies.
SS It was interesting to me to hear three different people bring this up. With Michael you could understand that it had a definite impact. Both Steve and Don also mentioned it. So in the early '70's in the beginning, it was really a crystallography problem without any biology. I want to ask you when you began to think about the biology of the system?
JJ It was years later. I was fascinated by quasi-equivalence. I loved symmetry. I loved organizational principles of molecules and I was fascinated by quasi-equivalence. I was absolutely just very, very excited about how you could take the same gene product and it would assemble into pentamers some of the time and hexamers at other times, and they knew exactly when they were supposed to be which, in order to form these quasi-equivalent capsids. In an intellectual sense, I was interested in that aspect of it from the very beginning but the thought of actually being able to answer the question - I was more fascinated with the question than with the thought that I would be able to answer it in the short term. I wasn't really driven by that. I was fascinated by the crystallography.
SS Did you have any questions or arguments with the Caspar-Klug quasi- equivalence model? Was it something you agreed with or understood?
JJ I understood it only marginally in the first period of time that I was in the lab. I understood what the conundrum was and was fascinated by that. I was so fortunate and this is the important thing to mention here because in the early part - it was probably either in late '73 or early '74, I think early 1974 - Don Caspar invited me to come to their laboratory to learn how to use the Franks double focusing mirror system that Steve Harrison, when he was a graduate student, had actually applied to be able to resolve diffraction from very large unit cells. I spent a week in Don's lab; it was just a life-changing experience for me, partly just being around Don and hearing first hand all the ideas of quasi-symmetry. And then Don views structural biology from a different perspective than a lot of crystallographers do because he's interested in, not so much the atomic detail of how things work, but in the larger scale picture. He helped me do solution X-ray scattering studies that have proven to be very interesting. So it broadened my perspective immensely and also allowed me to learn how to use this mirror system. Walter Phillips was a very senior person in Don's lab and Walter spent hours and hours with me. After coming back from Brandeis, we essentially "three-dimensionally xeroxed" the whole system back at Purdue and this was a huge turning point for us because we could get really good diffraction patterns from these crystals after that.
SS Was Steve still in Don's lab when you were there?
JJ No, Steve was at Harvard. Don had already moved to Brandeis then. This was perhaps two years after that whole group had moved from Children's Hospital to Brandeis.
SS You also mentioned that you didn't have rotating anodes. When did they come into use?
JJ When I came to Purdue, there was a rotating anode in the lab and it was in principle shared by Struther Arnott's and Michael's group but it wasn't functioning. It was an Elliot rotating anode and these were very, very unreliable instruments, but after I had worked with the still photographs from sealed tubes for awhile, it was absolutely apparent that we had to get the rotating anode generator running. I think it was probably in early '73, Michael brought one of the Elliot technicians from England and basically we spent almost a week together and rebuilt the instrument, got it running and in the process I learned the nitty-gritty of rotating anode generators. I would say for the next probably 4 or 5 years, I was the person that was primarily responsible for maintaining this instrument and I spent a very large amount of my time, maybe up to a third of my time, doing nothing but maintaining and keeping this thing running. I remember being questioned seriously by other postdocs in the lab as to whether this was a good investment of my time and that this was a technical problem that should be dealt with by technicians. I felt the end justified the means and tried to rationalize and explain my position whenever this came up, and it came up frequently. But ultimately we got better and better performance and I think probably because I understood the fundamentals of things we were able to improve the operation of these instruments. I stopped doing it seriously in the early '80's when we hired a technician to do it, but they were very temperamental pieces of equipment.
SS Before we go to the biology, I thought I would just let you tell me some of the interesting memories you have of your experiences in Michael's lab - not necessarily the science.
JJ It was an interesting time for me personally because Michael was totally consumed by the structure of glyderaldehyde-3-phosphate dehydrogenase and its relationship to LDH. It was in '74 or '75 when Michael put forward his ideas about so-called super secondary structures; finding common motifs in different protein structures and the Rossmann fold was the outcome of this analysis. I think this was really tremendously important because in the early '70's, people were not expecting to see thematic behavior of protein structure. Comparing LDH and G3PD, it became immediately clear that there were highly conserved regions, so Michael was just very, very enthused about this and was not really too focused on the virus part of things because there was not much interest in it. This was terrific for me because I was able to push along at my own pace and also I was learning a lot about viruses from the biological side of things and it was nice because nobody else knew anything, so if you knew anything, you were considered the expert in the area and I managed to kind of move into this area of expertise.
SS Were you learning this on your own?
JJ Yes, on my own and with help from Andy Jackson and Richard Lister. The first postdoc that joined me full time to work on viruses was Toshio Akimoto from Japan. Toshio was a tireless worker and collected the first complete data set from southern bean mosaic virus which went to about 10 angstroms resolution.
When we got that first data set in 1976, we really got our first structure information. That was 4 years after I had come. This was a 22 angstrom structure that we published in Virology. But I remember how excited I was because at that time, electron microscopy studies of cowpea chlorotic mottle virus had been done to high enough resolution to clearly see the hexamer and pentamer clustering of the subunits. In the electron microscope, southern bean didn't look anything like that. It looked like a smooth sphere without any kind of delineation. In the 22 angstrom X-ray structure we could see very clearly that there was clustering comparable to what there was in CCMV. While I think nobody ever anticipated that there would be this common beta sandwich-fold showing up everywhere, that was my first realization, that maybe these viruses were going to be more similar to each other than we thought. It turns out the SBMV fills in with protein between the hexamers and pentamers and with negative stain you don't get the delineation of hexamers and pentamers. In the x-ray map, they were clear. So I remember even at that low resolution, I was absolutely thrilled and we were running around saying "wow, these viruses look similar to each other".
SS At that stage, Steve must have already published something about tomato bushy stunt virus?
JJ Not in '76.
SS He must have published a low resolution map.
JJ Right, I am trying to remember. They had a 5 angstrom structure at that time because I think it was at the Erice (Sicily) meeting in 1976 that he presented it.
SS Were you at that meeting?
JJ Yes I was at that meeting and we were kind of the caboose of this activity because Steve did have the 5 angstrom map. In Steve's case, they had these protruding domains that were forming dimers and I remember that there were two features of Steve's structure that they were really thrilled about. One was the hinging behavior of these dimer domains when they formed icosahedral dimers versus quasi-2 fold dimers. And then the other was the so-called extra-density that appeared at the icosahedral 2-fold axis which at that time - of course it was just density we had no sense of connectivity or anything - it was thought it might be nucleic acid. In some ways that was looking forward to things that we saw later where nucleic acid was actually playing a very similar role.
SS Much later!
JJ That was much later. But this time (with TBSV) it was clearly protein. I produced this 22 angstrom map. I remember very well because it was in the early spring of 1976, and I remember stacking up the transparencies that we had plotted the density on and it just looked like cowpea chlorotic mottle virus that we had seen in the micrographs. This really preceded the Erice meeting and I hadn't seen anything of Steve's map so I am sort of describing more of a personal excitement than what would be a corporate excitement.
SS Well, it's an interesting point because both Michael and Steve claim that the structure of southern bean mosaic virus was their first realization of how similar these viruses were and yet, in fact, they should have been primed for it based on what you're saying.
JJ Certainly in a quaternary structure sense, at that resolution they looked similar. At higher resolution, actually even in a quaternary structure sense, they really are not that similar. All of these viruses tend to gravitate towards the shape of geometric solids related to the icosahedron. SBMV and TBSV both are very nicely described by a solid called a rhombic triicontahedron and I remember Steve pointing that out at the very beginning. Whereas the structure of cowpea chlorotic mottle virus is better described as a truncated icosahedron and so it's very interesting how you get this very close tie of different geometric shapes all of which have icosahedral symmetry, but have a very distinct morphology and the viruses tend to gravitate into those. So my impression of the similarity between SBMV and CCMV was maybe in some ways a little bit overreaction because at high resolution they weren't that similar, but clearly the hexamer/pentamer clustering was.
SS Did anyone pay any attention to the 22 angstrom map?
JJ We published it in Virology. It was very well received, actually. The reviews were very positive, but I think people looked at it and thought - so what. (And Jack laughs.)
SS What happened next?
JJ We pushed to 5 angstrom resolution.
SS Did that require anything special?
JJ It required a tremendous amount of effort. My time was spent in three areas - my personal time. I was keeping the rotating anode running, trying to improve our ability to collect data from very large unit cells and programming. I was developing the so-called double sorting method for doing molecular averaging that was developed in parallel, but independently, by Gerard Bricogne. Gerard was developing the software that Steve would eventually use for bushy stunt and I was developing the software that we would use on southern bean mosaic virus. Philosophically, they were the same procedures but they were developed entirely independently and Gerard then went on to develop a whole package of programs that became quite widely used. I never had the vision, shall we say, to be able to put this into a generally useable package.
SS However, virologists probably don't know anything about Bricogne's contributions.
JJ Anyway, just to go back in terms of the events that took place, I can summarize them quite succinctly, I think. Two postdocs - Ivan Rayment and Dietrich Suck - joined the group in '76 or perhaps a little earlier. They made tremendous contributions. Ivan pushed the data collection to higher resolution. In fact, Ivan developed all of the procedures associated with the oscillation photography which we were using. Toshio collected his data using precession photography. It was an heroic effort and then he moved to oscillation photography. In parallel, Michael was developing the software for processing these patterns and being able to scale the data and so forth. But Ivan was making huge contributions in the production of crystals and data collection and Dietrich was working on software development in terms of heavy atom determination and using search functions and things like that. Both of these guys were just absolutely top of the line people that have gone on to have stellar careers in structural biology - neither of them have anything to do with virology anymore. But they were making tremendous contributions and then by, I think '78, we published a 5 angstrom structure. It didn't dawn on us how similar it was to bushy stunt. We still weren't prepared to think about that and then in the later '70's and early '80's, two really important figures entered the picture: Andrew Leslie and Tomitake Tsukihara. They were both very, very strong people and actually it was Andrew Leslie that eventually traced the chain.
By '78 I got an independent position at Purdue so in a sense I was out of the picture officially by the time the structure was eventually solved in late '79, I think it was. I was off trying to do my own thing, but I was still very much physically there and I know that Michael related all of the frustrations of the electron density not being fully interpretable even though from everything that we could see it should be.
I remember a particularly striking event that took place one day because we all spent hours and hours and hours staring at these electron density maps and not being able to follow the connectivity of the chain. We were all leaning on this very large light box and suddenly the entire glass surface collapsed and it crashed down onto the florescent bulbs with much excitement, of course on the part of all of us. I remember Michael just looking utterly and completely frustrated and walking away from this scene without saying anything. We're all sitting there looking at these broken light bulbs and glass and the electron density and I thought to myself - um, this kind of characterized where we were in this project and I thought psychologically it's very important that we get this light box put back together again as fast as possible. I ran down to the shop and explained to the guys what had happened and what we needed - and I really wanted it to be plexiglass now instead of glass, so it wouldn't crash again. They understood immediately the gravity of the situation and literally within about 12 hours we were putting a brand new piece of plexiglass on the light box and we had all the large florescent tubes to go on there and we were back looking at the map again. I felt like the project had been symbolically resurrected from the catastrophe of the light box.
Michael's story explains exactly what had happened with the electron density maps although I played a part in this because the suspicion was in the programs that were doing the averaging and he was convinced, I know, that there were bugs in these programs. I kept coming up with control experiments to demonstrate that the programs were working O.K. and that it wasn't my fault. I know that up until finally when Tsukihara figured out that we had the wrong lattice constants, he thought there was something wrong with my programs. So I was greatly relieved when that got off my shoulders. The other part of this that I remember very, very dramatically was when Andrew Leslie had traced the chain and Andrew made the announcement basically, I'm sure Michael was involved too, but I remember coming in as kind of an outsider at that point and hearing that the fold was exactly comparable to bushy stunt and we all sort of just stood there in awe and were astonished because it was completely unexpected.
SS That's what Michael said and I guess Steve did too - that no one really had expected it to be so similar.
JJ And this was so hard to believe for people in today's mind set where every protein that's solved must have an analog that's already been around some place. It's very hard for people that have been raised in this comparative mind set to realize that there was a time in structural biology when we were all sort of expecting everything to look different from each other. It seems pretty na•ve now, but when you look back -
SS Michael and I discussed the surprise when rhino and polio were so similar and I had to bring myself back to where Michael and Jim (Hogle) were at that time because it seemed to me obvious that if the plant viruses were that similar, that the picornaviruses were likely to be similar.
JJ And then the animal viruses have these folds that are so, so similar to what we had seen in the plant viruses. It was just an astonishing set of revelations that were coming forward. It was really a hugely exciting time by the time we got into the mid-80's.
SS I wanted to ask you and maybe this will lead you into other things. I don't remember whether there was much you that could say from the 5.5 angstrom map about the biology.
JJ There was very little. That map, that paper was really more of a technical paper, I think, that just represented the fact that we could find heavy atom sites and that it behaved properly. We published it in Virology. There were 3 papers that I recall that we published along the way. We had a 22 angstrom paper, we had an 11 angstrom paper that was published in Acta Crystallogr. and was again very, very technical in nature, and then the 5 angstrom paper that was published in Virology again and I would say in some ways that the 22 angstrom paper made the greatest contribution to the biology in the sense that we had a little bit of a revelation there (the similarity to cowpea chlorotic mottle virus). The thing that I remember most about the 5 angstrom paper was that it was very clear that there was this helical cluster around the quasi-3 fold axis and you could see that very nicely and, of course, we completely lacked any of these extended surface features that had been seen in bushy stunt and at that time we never thought about it - we just thought that there are two different viruses, there are two different subunits.
SS Let's go to the final structure then. When Steve published his paper and we talked about it, he said that there were two revelations for him. One was that proteins had arms, and the other had to do with RNA. He said he rethought his ideas about RNA and seemed to feel that from the point of view of evolution, it was better for RNA not to have such a rigid structure because there were going to be lots of mutations and, if there were too many changes, the particle would be dead. I wonder when you think about the southern bean mosaic structure and even the later work, if you wanted to address those issues.
JJ Well, I shared the excitement that Steve felt on how molecular switching took place, that in fact we had these N-terminal regions that could be ordered or disordered and they were actually a major determinant in the geometry of the subunit contacts. So, in a way, this was the answer to the question that I had asked in 1972: how do the same gene products change the way they relate to each other? And the answer is an order-disorder phenomenon associated with arms. Of course it still doesn't answer how it happens. We always look at the finished product and in my dreams I imagine the dynamics of how do these proteins know where they are at doing this assembling process and so forth and so on. This still remains for me one of the real exciting things available to look at and understand - how is this taking place in a dynamic sense? But it certainly answered the basic question: this was the mechanism for how it was done and that was very exciting.
I remember being really impressed with the subunit interfaces because this was one of the first times where we had a large number of subunit interfaces. Obviously, there had been a lot of oligomeric proteins solved but I really had never paid much attention to them. But I was totally fascinated by the role of metal ions mediating subunit interactions and I remember looking at these for hours and hours and days and days and being so impressed with the difference in the nature of the subunit contacts. When you looked at subunits related by quasi-3 fold symmetry, these were really quite hydrophilic, ionic, with metal ions participating in mediating those interactions. When you looked at the contacts along the dimer axis, these were much more hydrophobic, much more flexible because they had to hinge. The subunits can exist in these two forms and I was really excited about all that and most of that probably reflected my naiveté because I had never studied protein structure before. This was the first time I really studied protein structure: when it was our structure! I really hadn't thought at all about the nucleic acid. We all hoped we would see something in those viruses. We saw nothing; it was completely disordered at the resolution we were working.
SS I have one more question to ask you about Michael and we might as well do it now and then we can go on to talk about your own lab. Michael has a whole other history because of his early childhood, first in Germany and then when he left Germany with his Mother. I wondered, since you're from the Midwest and somewhat isolated from Europe, being in Michael's lab did his background and experience with Nazi Germany have any influence on you at all? Did it make you aware of things you hadn't known about?
JJ Michael never volunteered information about that but sometimes we traveled together and we had opportunities for long periods of time together where we couldn't do anything but talk. I was totally na•ve and knew nothing about his background. But he would relate to me the experiences that he had, particularly as a child, and the terror he lived in when they were in Germany. He had some terribly difficult experiences.
SS I was asking more about how it influenced you because I assumed you knew nothing about that period.
JJ Probably, Michael was the only person I had personally been associated with that was exposed to the horrors of Germany at that time. It engendered a tremendous sense of respect for him. It also helped me understand him as a human being, because when you have a childhood under those kinds of conditions, basically it seems to me two things happen to a person. They either give up or they become an extraordinary fighter and I think we all know which choice Michael made. I think sometimes his behavior is frustrating to some people and, I think, understanding Michael the way I do, I rationalize that these are things that people develop. This doesn't relate to Michael but everybody has their strengths and their weaknesses. Sometimes I think if a person could just get rid of this or that weakness, they would be a very attractive individual and this would be so much better. But in fact all of us are this intimate mix and you can't possibly take away one set of characteristics and still end up with the positive so you end up with this remarkable blend of positives and sometimes negatives. Partly being around a complex individual like Michael has made me a student of human beings and looking at this remarkable blend of characteristics that create great scientific individuals and just trying to appreciate all that goes into this. I think being around Michael has had a huge influence on me in that regard and in many other ways. I was incredibly fortunate to have spent more than 20 years with him.
SS I was particularly focusing on that particular moment because many Americans, even people like myself who are Jewish, were not so cognizant of what went on in Germany during the war and then being brought face to face with somebody who had to deal with it really changes one's perspectives on the Germany of the 30's and 40's and I would have thought that would have happened to you.
JJ It had a large impact on me in understanding the reality of that time, but it had an even bigger impact on me in terms of realizing all that goes into making each of us who we are. That was something I dealt with every day.
SS When did you move into an independent position?
JJ In 1978, January of '78. I joined the faculty as any other tenure-track faculty member.
SS Before you did that, did you think at all about leaving?
JJ There wasn't any place to go. I was trained in a truly novel area of structural biology long before anybody thought about having virus crystallographers. Only partly jokingly I said that I never made the first decision in my life until the late '80's because every other step that I took there was only one possible direction to go. (A comment made with considerable chuckling.)
SS At the time you were making the switch, did you also have to think about whether you wanted to continue in virology or was that a case where you didn't even think about it?
JJ I didn't even think about it. I loved virology. I loved virus crystallography. I knew that if there was anyway I could possibly do it, I wanted to do it for the rest of my life. I found these particles to be so fascinating.
SS And how about which one you would work on?
JJ I chose cowpea mosaic virus because it was a plant virus that we could get in gram quantities which was absolutely crucial at that time.
SS This was 1978 so this was before people would even consider something more sophisticated.
JJ Yes, that's right. What I loved about cowpea mosaic virus was that, in its biological behavior, it was so much like a picornavirus. The picornaviruses had been characterized as having a genome associated protein at the 5' end. Cowpea mosaic virus had that. Cowpea mosaic virus was polyadenylated, it was one of the few plant viruses that was polyadenylated, most of them had a tRNA structure at their 3' end. The capsid was distinctly different from anything we had seen before in plant viruses. Also instead of being formed by a single subunit type, there were two subunit types that were in it. I had rationalized it as being similar to other plant viruses but with maybe gene duplication or triplication of the individual subunits that were of a single gene product type in southern bean mosaic virus and I thought this was the kind of ultimate structure to look at and that's why we started.
SS Who were we?
JJ Myself and an undergraduate student. Then right after I joined the Faculty, Michael gave me some very good advice. He said you should go to a lab where they work on this virus so you can get into the culture of it. George Bruening was at the University of California at Davis, worked with cowpea mosaic virus and had space in his lab for me.
Michael suggested that I do this and it was terrific advice but it was an awkward situation for me because we had two very small children; a 3 year old and a 1 year old, and my wife had many reasons why it was too awkward to go away for 4 or 5 months. Michael was adamant that I shouldn't leave my family and that we should figure out some way to do this together. He said: "decide you are going to do it and everything will fall into place". That's advice that I have always stood by for the rest of my life after that because I think it's so true if you make a decision you are going to do something, then everything else will fall into place, and it did. I spent four spectacular months with George Bruening - from May to August of '78. We became close friends; we remain very close friends to this day. And it got me into the virology community because we still hadn't broken in there. In your early discussions with Steve and Michael it was clear that the structural part of this had not been fully appreciated. George was a physical chemist and he really liked structure.
SS What research was he doing at that time?
JJ He was sequencing the genome of cowpea mosaic virus and was characterizing the properties of different mutants and there were a lot of interesting little behaviors of cowpea mosaic virus that he was characterizing. So when I came back from Davis, I really had learned a lot about cowpea mosaic virus. I had a contact and I could pick up the phone and talk to him about anything. Joel White was the undergraduate student and Joel had produced the first crystals while I was in Davis. We published a little paper in Virology on the cubic and hexagonal crystal forms and we started trying to collect data.
SS What happened to him?
JJ Joel got a B.S. from Purdue, then went to Ohio State and got a dental degree. Then he went to the University of Michigan and got a Ph.D. in biomaterials. He's been on the faculty of University of California, San Francisco for a number of years and continuing work in biomaterials, so that worked out beautifully.
SS How well did these crystals diffract?
JJ To 3 angstroms resolution. They diffracted well. I think we were learning more and more how to grow crystals. Most of these viruses will eventually behave if you are patient enough to look at all of their crystallization properties. Joel did a wonderful job on cowpea mosaic virus. There was a tremendously interesting little anecdote that goes with this because the two forms that we had were a hexagonal form that had a l000 angstrom lattice constant and a cubic form that was actually very similar to the crystals of bushy stunt; body centered cubic with lattice constants of around 320 angstroms. We could grow the hexagonal form very easily and reproducibly but the cubic form was very difficult to produce. So the question was: do you work on something that everybody thought was impossible to solve, something with a 1000 angstrom lattice constant or do you forget that and just work on the one and just keep trying to get these crystals? Since I knew how to do crystallography and was much more comfortable with crystallography than with biochemistry and all that part, I started to do as much as we could with these hexagonal crystals. That was really a good idea, because, while it wasn't until 3 years ago I think that we solved the structure of the hexagonal form, it introduced me to a lot of things.
We couldn't collect data on a rotating anode. We had to use the synchrotron so I was the first person at Purdue to start looking at synchrotrons. Hal Wycoff, then at Yale, was very enthused about a major synchrotron effort in the United States and he was the pusher for the light source at Brookhaven. I remember that Michael wasn't too enthused about this because he liked working in his laboratory and so forth. But I went to Stanford (SLAC) and had a disastrous trip out there trying to collect data. Then I went to LURE in Orsay, France. This was one of these moments that kind of defines what it's all about because Paul Sigler had collected data there on a crystal that had a 500 angstrom lattice and we came in with a 1000 angstrom lattice. I was by myself, we couldn't afford to have more than one person go.
SS Did you have to work 24 hours a day?
JJ No, the French are civilized. The synchrotron ran only 18 hours a day! We put our crystal on. I was working with Roger Fourme. The French were very helpful, very fine people and they gave me a lot of assistance. I will always remember this. We took this exposure - we had taken stills and it was clear that it would take a pretty long exposure because we had cut the beam down until it was quite small. Anyway, I went to the darkroom and pulled the film out of the fixative and there were just spots everywhere and they were individual distinct diffraction measurements, obviously measurable and I remember:
I totally lost control. I was just literally dancing around the floor of the synchrotron waving this picture.
SS I hope the other people there could appreciate it.
JJ Well, Roger certainly did and there were some others. But anyway it was just one of the most wonderful experiences I ever had and this fired me up for years, literally.
SS You were the first person then really to realize the power of the synchrotron.
JJ Well, certainly at Purdue
SS But Michael was given credit for the first person to do virus, because he did rhinovirus on a synchrotron.
JJ Right. But in terms of the virus crystallography community, I probably was the first person that did these experiments. I don't remember whether Steve had done any experiments with the synchrotron. This was in 1980, late '80 or early '81. Stanford and LURE were the two places where this was taking place and then not long after that, Cornell started developing a very strong program in structural biology at CHESS with Keith Moffat being the driving force. At Purdue, we used CHESS exclusively for many years.
To go back to CPMV for a minute, eventually we were able to get the cubic crystals again and it was an undergraduate student again that got these. Another one of these little anecdotes. I had told the student, Melissa Harrington, to grow crystals in a certain way. She came to me and said that the crystals look very funny, and when I looked I saw that there was a whole dish full of cubic crystals. I remember having this feeling that I must be very, very careful to get all of the information. I stood up from the microscope and said: "Melissa, now come back to my office with me". We sat down and I said: "I want to know exactly what you did". She explained and it turned out that she had used a phosphate buffer and we actually hadn't been using any buffer at all, and that was the key. So as long as you had phosphate around you got cubic crystals.
Then that launched that project and I was joined by two post-doctorals. One was Ramakrisha Usha who came to me from Bangalore. She worked very diligently and found the heavy atom derivatives that allowed us to solve the structure. The other, Cynthia Stauffacher, was actually the one who traced the chain.
There was a meeting, I think in 1985, outside of Strasbourg. It was a structural biology meeting organized by Dino Moras and Dietrich Suck. It was really remarkable because Jim (Hogle) was there with polio and Michael (Rossmann) with rhino. We were in the "caboose", coming along with a proposed tracing of cowpea mosaic virus. But by that time it was absolutely clear that cowpea mosaic virus was very much related to the picornaviruses structurally as well as in terms of the other details I spoke of before. We didn't have a really good chain tracing, but the proceedings of that book was the first place where we published the structure of cowpea mosaic virus. So this joined the plant viruses in yet another way into the evolutionary development with the mammalian viruses and of course we were really thrilled.
SS Were there other insights that you could get from these structures beside the evolutionary ones?
JJ Yes, by having all three of these structures - the two mammalian viruses and then our plant virus - at the same time one could see that the mammalian viruses have these very elaborate surfaces with all these antigenic sites and canyons and so forth and so on. All of these were created essentially by insertions between strands of the beta sandwich. They looked very spherical and at high resolution extremely complex. When you compare them with the plant virus, the plant virus had a very distinct morphology, even in the electron microscope and it didn't look like the picornaviruses at all. The reason for that is these plant viruses have none of these elaborations. The canyon's gone and the whole south side of the canyon has disappeared; all of those inserts that create the so called south side of the canyon are gone. There are just tight turns between strands and all of the antigenic sites are again very tight turns. So it was absolutely clear that the animal virus has many more things to deal with, both in terms of receptor binding and in terms of avoiding the immune system, setting up these decoys and things. The plant virus is just a streamlined version. If you don't need any extra loops, you don't put them there. Again, it is an evolutionary issue. The comparisons made it very, very clear that the plant virus is living in a much more simple environment.
In terms of the basic structure we didn't get too much else from cowpea mosaic virus, but not long after CPMV, we solved the structure of bean pod mottle virus which was a relative of cowpea mosaic virus and that turned out to be the first time that I wasn't in the caboose! We actually got in the engine on that one because it was the first time that we saw extended regions of ordered nucleic acid in a virus. They formed these magnificent trefoil structures around the 3-fold symmetry.
SS Was the resolution so good that you could tell that you were looking at a base?
JJ You could see the bases, you could see the ribose phosphate backbone.
SS I can imagine that at first you didn't know what you were looking at.
JJ That's happened more than once!
SS I am trying to imagine what it was like when you were thinking you were looking at amino acids.
JJ The driving force on that project was Zhongguo Chen. He was a sabbatical visitor who had originally come from Peking University and the idea was to spend a year and then go back. This was in 1989 and he and his wife decided they were not going back.
SS That was a decision after Tiananmen Square?.
JJ Right. Peking University is probably one of the more liberal universities in China and probably Zhongguo was among the liberal of the liberal and they could not face going back. So he stayed on. It was horribly traumatic for them, but terrific for me. He's gone on to be a research director at Merck now so it worked out fine in the long run. But anyway Zhongguo immediately recognized what he was looking at. I'll always remember this - it was such an exciting day. All of our graphic machines were in one room at Purdue. Zhongguo came down and said: "you have got to come and look at this". I said: "sure". So I went down and I felt like we were at the foot of the Tower of Babel. It was a very international group of people and we were all looking at this and everybody seemed to start speaking only in their own language. The Indians were talking to each other and the Chinese - whoever they could talk to in their own language. It was just one of those moments. We had never seen it before and it was just such an exciting result. This was published as an article in Science - sheer serendipity with nothing to do with one's skills. It just happened, and I always said I would rather be lucky than smart any day.
SS Let's go back to cowpea mosaic virus before we get off on other things. You had mentioned earlier that you had been interacting with George (Bruening). Was he able to use the information that you were generating? Was there any way that you two could interact?
JJ George was very interested. This virus undergoes a natural cleavage phenomenon. The C-terminal of the small subunits that are clustered around the 5-fold axis get processed and you end up with a heterogeneous population of particles. Since there was a lysine residue on the end of this you could separate these things by whole virus electrophoresis and you get these two nice species of particles, all cleaved or all uncleaved.
SS Did you have to do that for the crystallization?
JJ No, because George had a mutant that spontaneously cleaved all the way. For our crystallography, we used this Bi1 mutant that he gave us.
SS That's important!
JJ Terrifically important because in 1978 when I arrived in Bruening's lab we were talking about homogeneity and so forth and so on and George said: "ah, I've got something for you". And it was shortly after we sent a preparation to Joel White, back at Purdue, that we started getting the crystals. To this day we use the Bi1 mutant of CPMV for all of our crystallographic studies.
Other things, let's see. These particles existed as an empty capsid and as nucleoprotein components. Actually there were two nucleoprotein components because the RNA genome was split; so you have two pieces of RNA and they were packaged separately. On a sucrose gradient you see three peaks that were separated by the amount of RNA that was in them. We also solved the structure of the empty capsids.
SS You had to separate the different capsids before you did the crystallization.
JJ Right, we usually did it on a cesium gradient because you could load a lot of material on a cesium gradient and we got really nice separations. But when we solved the structure of the empty capsids, we saw the influence that RNA had on the order of the capsid because, again, we had an arm, the N-terminus. When there's no RNA it was completely disordered and we didn't see it in the electron density at all. These were subtle things. I don't think there were any breathtaking results.
SS But actually, and this is now more related to things I was interested in and that is the ability to form empty capsids, was this because after cleavage there weren't so many positive charges?
JJ No, the empties just formed naturally. The amount of empties actually depended on the growth conditions of the plants. If you grew them at lower temperatures, you would generate significant amounts, maybe up to 25% of the total virus were empties. Under warm conditions in the summer, it dropped down to 5 %. But that's just what came out of the plant, there was nothing by design there at all. And I don't think it was related at all to the cleavage at the C-terminus. Getting back to that cleavage for a minute, it was interesting because it was clear that the C-terminus was completely exposed and the last residue we saw, as I recall, was a leucine residue. It rationalized nicely that this was something that could be cleaved very easily, but it didn't contribute too much to the excitement.
SS You already talked about what happened in '89 (seeing RNA in the structure). Had you started with nodavirus by then? I don't want to jump ahead too quickly.
JJ We started with nodaviruses in '83. Roland (Rueckert) came down (from Madison to Purdue). He and Michael had been collaborating, of course, for years at that time. I can maybe make one contribution about the development of the picornaviruses because that was a nice little anecdote.
I met Roland in 1978. I went to one of the NATO meetings on virus structure. It was right after I had been in George Bruening's lab and this was held in Maratea, Italy. Roland was there, a lot of different picornavirus people were there. It was a 10-day meeting, very casual, a lot of opportunities for talking. Roland and I talked extensively and I told him my hopes and dreams with cowpea mosaic virus. Fred Brown was there. Fred was wildly enthusiastic about the similarity between animal and plant viruses. Roland invited me to Madison after Maratea and we had a nice relationship. I'm not sure whether Roland and Michael knew each other prior to the Virology conference in Strasbourg in 1981. I remember vividly that at that time, Michael was really thinking pretty seriously about poliovirus as being what he wanted to pursue. I remember having dinner with Roland one night in Strasbourg and Roland saying: "you know, Michael should really work on rhinovirus". It didn't make sense to him for Michael to compete with Jim (Hogle) because it was always clear that Jim had gotten the first crystals when he was still with Steve. I said: "you should really talk to Michael about that because maybe that would have an influence". The next night Michael, Roland and I had dinner together in Strasbourg and Roland laid out all the reasons why he thought rhinovirus was at least as interesting as polio and that he was willing to bite the bullet and produce the first quantities of material necessary to get the crystallography going.
I don't know whether Michael recalls it the same way as I do or not, but I'm pretty sure that it was at that meeting in Strasbourg where the real decision was made. That was in the summer, and then I remember vividly John Erickson, Michael and I driving up to Madison to meet with Roland. That was when I started getting associated with Paul Kaesberg also and we had the first Wisconsin-Purdue meeting, the Wispur meetings. That was basically the four of us. I can't remember other people from Roland's lab that were involved but they laid out all of the plans there and John Erickson went and spent a period of time with Roland learning how to do the preparation of rhino and that was the start of the rhino project according to my memory.
SS Several people have told me the same thing.
JJ That Strasbourg meeting was crucial in getting all of this going and the whole history could have taken a very different direction if things hadn't connected at that point in time.
SS You were starting to tell me how you got involved in nodavirus.
JJ Right. Rhino was being solved. You know about the marvelous story of the rationalization of antigenic sites and canyons and all of these kinds of things. Roland also had a very strong interest in these little insect viruses, the nodaviruses. During one of his visits he came in and threw an electron micrograph down that had been taken of a drosophila cell and it just had this sheet of beautiful crystalline virus particles in there. He said: "this was black beetle virus", and asked what I thought. Well, I thought it looked really, really good.
SS So he decided to come to you instead of going to Michael?
JJ I think with Michael, the foreseeable future was filled with activities associated with the drug-binding. I don't think this was something that Michael probably had any significant interest in. Actually I'm jumping the gun on this, I think that was probably in '83. I don't think they had solved the structure yet. Michael was completely occupied with rhino and this would have been a distraction. But it wasn't for me, because we were ready to go. Again I just had such marvelous good fortune because there was a postdoc in Sunderlingham's group in Madison named Hosur who was coming to my group. He was working with Roland at night learning how to do all the tissue culture work with black beetle virus. Hosur came down to Purdue then and we set up drosophila cell culture for the first time and it was all seamless and transparent because he had been working in Roland's lab for a number of months, late at night. Sunderlingham was complaining to me because he thought I was influencing his postdocs in a negative way.
Hosur did this wonderful work on black beetle virus. When that structure was solved in '86, we didn't recognize RNA when we saw it because we had this piece of density that was sitting in a comparable place where the extra protein density for bushy stunt was sitting, right along the twofold symmetry axes, the line at the twofold related subunits and they were parts that we couldn't account for in the protein structure so we assumed that the "island" of density was formed by disorder and then there was some order (the island) and then it was disorder. We called it the arch peptide because it formed this beautiful helical arch. It was only later when Andy Fisher was in the laboratory and solved the structure of flock house virus which is very, very similar to black beetle virus, 85 %sequence identity in the capsid protein, that we could see base density. This was another experience down in the graphics room because it turned out that it formed this gorgeous duplex and right near the 2-fold symmetry axes.
SS You didn't have to break a light-box.
JJ No, no broken light boxes, just shear delight.
SS Let's go back, when you looked at the density, you said that you couldn't tell it was nucleic acid, did you just assume that it was disordered and that's why you couldn't trace any chains?
JJ We just assumed that it was protein. We didn't see any bases at all. All we could see was the phosphate backbone which we just thought was the C-alpha backbone essentially and that the side chains were all disordered. After the fact, it formed a perfect helix, but the other RNA density we saw wasn't helical and we were just weren't prepared for it.
SS In the recent work that we've been talking about it seems like things went completely smoothly in terms of determining the structure. You don't mention any problems with methods. By this time were you using the synchrotron? Were there other problems that you had to solve? It seems as though you went from a period where the barriers were the crystallography and programs to now where that was routine. Is that true?
JJ All of the postdocs working with me were highly trained crystallographers. Everybody that came into the lab had a reputation already as a crystallographer, certainly not as a virus crystallographer, but some as protein crystallographers, some as small molecule crystallographers. All I ever hired as far as postdocs were concerned were always crystallographers and that held really up until the mid-'90's. There were problems; but the major problems that we had encountered on southern bean were utterly inadequate computing. Historically, viewing it from now, we were just crazy to try to do the things that we were doing. An electron density map was held on 12 reel to reel magnetic tapes and to read 12 magnetic tapes without parity errors was almost unheard of. You just kept reading. We knew that there were errors during the reading but we just did it anyway. It was basically data collection and data processing and computing. Those were the big problems. The synchrotron, starting in the early '80's for me, dramatically reduced the amount of trouble there. In 1983, we got the Cyber205 Computer at Purdue. When we were doing southern bean mosaic virus, it took us 6 weeks - 6 weeks - to do one cycle of molecular averaging and Fourier transformation to produce a new set of phases from this averaging procedure. When we got the Cyber205, I spent all of my time for about 18 months rewriting all of the programs to run on a parallel processing machine. When everything was up and working, which took about 18 months from the time we got the computer, we could do 6 cycles in one day. It was just breathtaking. In my mind, sure, there were problems, there were difficulties but compared with what we were trying to do with southern bean mosaic virus, everything seemed much easier.
SS So when you had people who were just crystallographers, did they become biologists? How did you, yourself, make that transition? This seems particularly important as we start talking about nodavirus because there is much more biology with these viruses. One of the contributions that Steve's lab made (with plant viruses) that had a big influence on my work was his studies on disassembly. During the time that you were at Purdue, were you able to branch out at all into more biological kind of questions?
JJ I wanted to, but we never did anything that was useful. We did a lot of different kinds of experiments on CPMV, but mostly they kind of stimulated us. I don't think I ever published anything on the biophysical studies that we were trying to do on CPMV.
SS How about other plant virologists?
JJ There was not a lot of reaction. I think even with George Bruening, it was probably the warmth of our personal relationship that kept us together rather than any kind of substantial scientific collaborations. In fact I don't think I ever published a paper with George.
SS Do you have any idea why plant virologists weren't picking up on the structural work?
JJ As I recall, there were really only two people that were very enthused although I'm probably not being fair on this. I remember Roger Hull from the John Innes Institute being very, very excited about the structure and really asking lots of questions; he was a biophysical type of guy. Then John Bancroft was also very interested. He was interested in assembly. I don't remember very many people having a lot of interest. I don't even know whether there is much interest today.
SS I know Mark Young has been interested, but that must have begun when he came to Purdue.
JJ Mark Young, when he came to Purdue thought: "I am with all these structure people, I am going to learn structure". And of course we had just an absolutely marvelous time with Mark. But I don't think he had a strong interest in it before he came to Purdue. I think it was the circumstances and Mark's enormously energetic and opportunistic attitude. He didn't learn crystallography, as such, but he learned structural stuff very quickly. I think it's exposure. I don't think we've managed to get enough plant virologists interested. Another thing is, let's face it, plant viruses just aren't as interesting as animal viruses. We're completely excited about nodaviruses because they do so many interesting things. They have autocatalytic reactions going on. The animal virus just has a lot more interesting problems to solve than the plant viruses does.
SS Jack, you were going to say something more about Maratea.
JJ Well, I mentioned before that that was where I met Roland and that initiated our relationship which then eventually spawned into the relationship between Michael and Roland on rhinovirus. But the Maratea meeting for me was of enormous importance because I got to meet so many people in the polio and picornavirus field. I just remember vividly the influence that Fred Brown had. It was difficult for me because I didn't know anybody at this meeting. I had never met anybody there before and I remember Fred coming over and sitting down as I was sitting eating by myself and engaging me in conversation. I told him what I was working on and he wanted to see my poster and he expressed a lot of interest. Then he introduced me to Roland and Ellie Ehrenfeld and Ellie's husband, Don (Summers), and he just kind of integrated me into the whole group.
SS When was this meeting?
JJ 1978
SS By that time had you become more familiar with the biology of viruses?
JJ I was very excited about the biology. I was reading everything I could read about picornaviruses because of the analogy with the plant viruses that we were working on. I was really excited about the biology of these systems and of course this meeting was almost all biology, there wasn't any structure. Nobody had crystals of any picornaviruses. Primarily, it was establishing all these relationships (between scientists) that exist to this day. I think, scientifically, it is so important to have these kinds of relationships because they keep your compass straight. People tell you before you publish whether they think it's useful and there's just a lot of encouragement when you are a young person and it's really vital. So that meeting really brought me into the world of the virology side of what I was up to.
SS Let's go back to nodaviruses, because I remember some of the excitement with nodaviruses in their being dynamic, but I don't remember how that came about.
JJ Well, like most of these things, there are sort of stages of realization and one of the things that we had learned early on - immediately from the structure was that the virus - - -
To go back, Roland had determined biochemically that the virus underwent a maturation cleavage and that the subunit assembles as a 407 amino acid polypeptide and then post-assembly, it cleaves between residue 363 and 364. This is autocatalytic, we assumed, because it is totally unexposed to the surface, it's inside the particle. Yet the fact that this maturation cleavage occurred, immediately implies that there were some dynamics associated with the polypeptide chain at this point because when it assembles it's connected and when it's cleaved, it's cleaved. There must be some level of dynamics associated with that. Then as we studied this more and more, we felt that this cleaved off polypeptide was probably involved in interacting with membranes and was potentially some kind of a conduit to get RNA across membranes. So the kind of model that we started working with was that it was like a fusion peptide but it didn't really fuse membranes.
SS When were you proposing this?
JJ This would have been in the early or mid-nineties. We had solved the structure, we had a couple of different structures. Actually, one of them - and this is a real important thing to bring out. We had begun to work with Tim Baker and cryoelectron microscopy. There were two viruses that we worked on a lot with cryoEM - three - the first was cowpea mosaic virus. I hope it is okay to "retro" a little because this was probably my third most exciting moment in science when Tim Baker and I started working on a project where we wanted to see if we could image monoclonal antibodies attached to the virus. Marc Van Regenmortel in Strasbourg had made monoclonal antibodies to cowpea mosaic virus. His postdoc who had made those antibodies was Claudine Porta and she came to postdoc with me. She was probably the first person to come to the lab that wasn't a crystallographer. She was really an immunologist.
Claudine and a technician in Tim's lab, Wanda Wang, spent the better part of a year developing the technique to be able to get tight enough binding of these antibodies, these monoclonals, to cowpea mosaic virus so we could really see highly decorated particles in the negative stained images. Wanda did the three-dimensional reconstruction of this complex. I had been out of town. Tim Baker's father had passed away and I knew that he was leaving to go to his father's funeral. But I had this urgent message waiting for me when I got home that I had to be in the lab by 7 o'clock the next morning because Tim was leaving at 7:30 but he had to see me before he went. I thought that there must be some great crisis. I got to the lab went into Tim's room and Wanda was there. Wanda never came in early and I sort of got suspicious that maybe this was an interesting scientific result. And lo and behold, Claudine was there and Claudine never came in before 9 o'clock. They put up on the graphic screen the reconstruction and there was this magnificent image of cowpea mosaic virus just gorgeously decorated with the antibodies. Then we could make a footprint and we could show exactly where the antibodies were binding. It was tremendously exciting and I think it was really the start of combining cryoEM and crystallography, and the beginning of things that Tom Smith eventually did with antibodies to rhino and then, of course, the receptor bound to rhino that Michael did. I think the contribution that we made got kind of lost again because it was a plant virus.
SS But eventually it will turn out that knowing the structure of plant viruses will be useful in this (the 21st) century. (I am referring to work that Jack has been involved with in which immunogens are coupled to the surface of plant viruses to be used as vaccines.)
JJ We did all this cryoEM work with Tim Baker and then we did cryoEM reconstructions with flock house virus. What was exciting is that even though the RNA is not well-ordered, with very low resolution, you still see the density for the RNA. You are looking at sort of average occupation so you get a very clear sense of how the protein and RNA are relating in a general way. Then when you build in the atomic model of the protein from the crystallography into the cryoEM reconstruction, you start to get a very different impression of the relationship between the nucleic acid and protein. One of the things that we noticed was that the gamma peptides - the parts that get cleaved off during autocatalysis in nodaviruses - were actually sitting in a beautiful little pocket. You didn't have that impression when you looked at it in the X-ray map but in the cryoEM map they are just separated from the outer surface essentially by a couple of loops of the protein. So even though they are internal, they are still very close to the surface. We started thinking that maybe these peptides can get out and are involved in breaching the membrane and in getting nucleic acid across.
That's where we were when we moved to Scripps. I was very fortunate that Anette Schneemann came to Scripps as an assistant professor at the same time as I arrived. She was a graduate student with Roland and we had worked closely together at that time on the structure and function of nodaviruses. She is an outstanding molecular virologist and has designed a variety of methods for analyzing structure-function relationships. The various mutants of flock house virus, as well as the expression-assembly system that she has developed, placed us in a wonderful position for collaborations with colleagues at Scripps.
We have been delightfully inundated with opportunities to use our viruses for different things. They are big enough that they are pushing the envelope for a lot of different technologies, but they are small enough that it's feasible. Among the people that came to us shortly after we got there was Charlie Brooks for computational studies and then Gary Siuzdak for mass spec studies. I didn't even know what you did with mass spec for biological molecules. Gary defined a very simple experiment. He said: "we'll just put the virus into the solution, we'll put in trypsin and we'll let it chew away for awhile and do a time course; then we'll do mass spectrometry at intervals and by knowing the sequence of the protein and knowing the specificity of the protease, by the masses of products we can tell exactly where the cleavages have taken place". I said: "oh, that sounds good", and I suggested that maybe we'll cleave loops on the surface of the protein and so forth and so on.
Gary did the experiment and brought the data back. He knew nothing about the structure, but said: "beautiful results, we are getting these cleavages here, here, and here". I looked at it and I was immediately skeptical because the places that were getting cleaved were internal in the X-ray structure and, in fact, the most susceptible cleavage sites were the gamma peptides. While I had put forward the proposal that they could be externalized, I wasn't prepared to believe it! I thought it would only happen when it bound to the receptor. I couldn't image that these things were fluctuating in and out of the particles in solution when they didn't have any influence of other biological interactions. But the results were very clear and I expressed my skepticism about this.
SS Was this done at room temperature or at 37 degrees?
JJ It was done at 20C, ambient temperatures. It was very hard to change temperatures very much because you are changing the characteristics of the protease as well. I gained this enormous respect for Gary because I told him that this was outrageous and he designed control after control. I was worried that we were looking at some little population of broken particles or something like that, but he put in internal standards and was able to demonstrate that we were actually looking at the whole population. At the end of the day, we had nothing to believe except that this was what was happening. I then became resensitized to Jim Hogle's early work that had evidence that the N-terminus of VP1 was transiently exposed in poliovirus. So we published this work. We submitted it to Nature Structural Biology and I was expecting a very rough ride on this. Instead, both referees applauded the work and I think we had done good enough control experiments that we had answered most of what the anticipated questions would be.
I also remember vividly a conversation that I had with Michael when I went back to Purdue and I told him about these data. One of the wonderful things about Michael is that he is absolutely transparent and you never have to wonder what he is thinking. One of the interesting characteristics is the angle between his neck and his shoulders and the more uncomfortable he is, the more this angle decreases till it approaches zero. In the course of this conversation, I noticed that this angle was approaching dangerously close to zero and finally he stopped me and said very succinctly: "Jack, you and I both know that this is rubbish". But, I think today he believes these results and is very enthusiastic about the dynamics of these virus particles. For me personally it totally changed my view. What it told me was that the crystallography was very, very valuable as far as it went, but we had to come up with other kinds of techniques to understand what all the implications were.
SS I was trying to think of other examples and of course what comes to mind right away that would have influenced your thinking would have been the flu hemagglutinin.
JJ Exactly, but I think the thing that I wasn't prepared for was that this sort of dynamics went on in an equilibrium sort of way and that this exposure was a continuous reversible process.
SS The reason I asked about the temperature was, I was wondering, if the virus was continually doing this why isn't it being inactivated all the time.
JJ Exactly and there is a conundrum about this. It's not clear how it is being protected but interestingly it is only after hours and hours are you actually proteolyzing any of the exposed parts. The outer surface of the particle has clearly evolved to be very, very stable and resistant to any kind of proteolytic activity. These gamma peptides don't cleave at a very high rate, but they are clearly the most susceptible in a time course type of study.
SS Does this lead to more biological experiments? Could one try to increase the rate of proteolysis?
JJ One of things Anette did was to make antibodies to the gamma peptide and showed that when you expose the virus particles to these antibodies, you drop the infectivity by almost a log. This is perfectly consistent with the idea that these particles are in equilibrium and this equilibrium is an important part of the infection process. That was a nice kind of follow up. Anette also made mutants of the virus that do not undergo the maturation cleavage. They can't be made in a virus infection, but can be made in the baculovirus expression system that she had developed. When there is no cleavage, the proteolytic rate of the gamma peptides is reduced. Covalent independence is clearly needed for these things to fluctuate in and out. When the wild type form is expressed with the baculovirus system, there is cleavage and the protein is even more susceptible to proteolysis than the authentic virus that contains the viral genome. The baculovirus expressed particle is packaging essentially cellular RNAs and tRNAs and things like that and that must affect the equilibrium. These results have profoundly influenced my personal view of how crystallography relates to the biology.
SS Am I correct that there is a known receptor for nodavirus?
JJ There is one and we can see it on a gel but we have yet to be able to isolate it in enough quantity to get enough sequence so we can really tell what it is as far as the gene is. Our real hope is to be able to look at susceptibility. There are just a whole series of things to do if we can get a functional part of the receptor so that we can do in vitro some of these experiments - it would be enormously exciting.
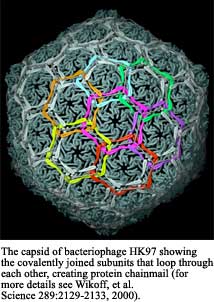
We are sort of "shooting the breeze" here and I can tell you that we just published the structure of the bacteriophage capsid HK97. This was tremendously exciting because here was a virus that naturally undergoes enormous dynamic changes in the course of its maturation. The subunit is completely unlike any other viral subunit we've ever seen and probably, the most interesting finding is that these capsid proteins form concatenating rings that interlock each other and make this sort of chain-mail.
SS Were there any interesting revelations there in terms of the moment of discovery?
JJ Yes, Roger Hendricks and Bob Duda had sorted out this whole story of the chain-mail. Once this virus is fully mature you cannot get it to go into an SDS gel regardless of how you treat it. The only way you can get it out of the loading well is to treat it with a protease. They came to the conclusion that these proteins would physically catenate with each other as well as chemically interact with each other and they knew the ligation site and so forth, but they envisioned something like two proteins doing a "dozy-do" as in a square dance where an arm would reach out and hook around. The structure reveals something that none of us had expected and that is that each set of hexameric proteins and pentameric proteins form a simple ring but they don't form the ring that we were expecting it to form. Whenever you look at the symmetry elements, quasi or icosahedral, of a capsid, you have subunits that are directly adjacent to that symmetry axis which usually form the morphological units of what shows up in the EM, but then you've got the set of proteins that are one step away. They are actually interacting with the proteins that are adjacent to the axis. So you have two levels of symmetry. You've got the hexamer directly around the axis and then you have a hexamer that's one step away from the axis and it turns out that if you crosslink the subunits that are one step away so that they just form a simple ring, the only way that you can keep icosahedral symmetry is if these rings interlock like chain-links. It's just a topological fact, if you relate these by two-fold symmetry. The easiest way to view it is if you have two hexamers and they are like this, there's a 2-fold symmetry axis in between them. The only way you can maintain the 2-fold symmetry is to have it go like that. We were absolutely stunned by this result. It's so beautiful! It's gorgeous! It was another one of those "holy cow" kinds of experiences.
 |
and |
 |
We are grateful to Bill Wikoff for providing us with "the real thing" and for the following photograph.
 |
| Introduction
| Some historical highlights: structural virology
and virology |
| Solving the Structure of Icosahedral Plant Viruses
| Picornavirus Structure | Poliovirus
| Polio
The Influenza Virus Hemagglutinin | The
Influenza Virus Neuraminidase
| Issues of Science and Society |
contributors| Home |